Introduction:
Cancer cell metastasis is a complex process that involves many different molecular mechanisms. One such process is the interaction between cancer cells and the extracellular matrix.
Quantitative gene expression analysis of Ti cells in response to cancer cell invasion done by using qPCR, RT-qPCR, and microarray technology.
The molecular mechanism behind the invasion of cancer cells into tissues sees through the visualization and analysis of gene expression in Ti cells.
Cancer is difficult to diagnose and treat. To understand the molecular mechanism behind cancer cell metastasis, researchers need to first understand the gene expression in tumor cells.
The team used a technique called microarray analysis to test gene expression levels in tumor cells and found that they are related to different stages of cancer cell metastasis.
Cancer is a disease that affects millions of people worldwide every year.
The team used a technique called microarray analysis to test gene expression levels in tumor cells and found that they are related to different stages of cancer cell metastasis.
Introduction: how can you track how cancer cells spread throughout your patient’s body?
There are many methods of tracking how cancer cells spread throughout your patient’s body. The most common method is to use of a PET scan, which is a type of nuclear medicine imaging procedure.
Introduction: The most common method of tracking cancer cell spread throughout the body is through a PET scan, which involves injecting small amounts of radioactively labeled glucose into your patient’s blood. This allows for the location and movement of cancer cells in the body tracked by looking at where the glucose is found on the PET scan image.
Introduction: There are many methods of tracking how cancer cells spread throughout your patient’s body. One popular method is to use a PET scan, which involves injecting small amounts of radioactively labeled glucose into your patient’s blood.
What is Spatial Transcriptomics?
Transcriptomics is the study of how genes expressed in a cell. It is the process of measuring changes in gene expression at a certain point in time and space.
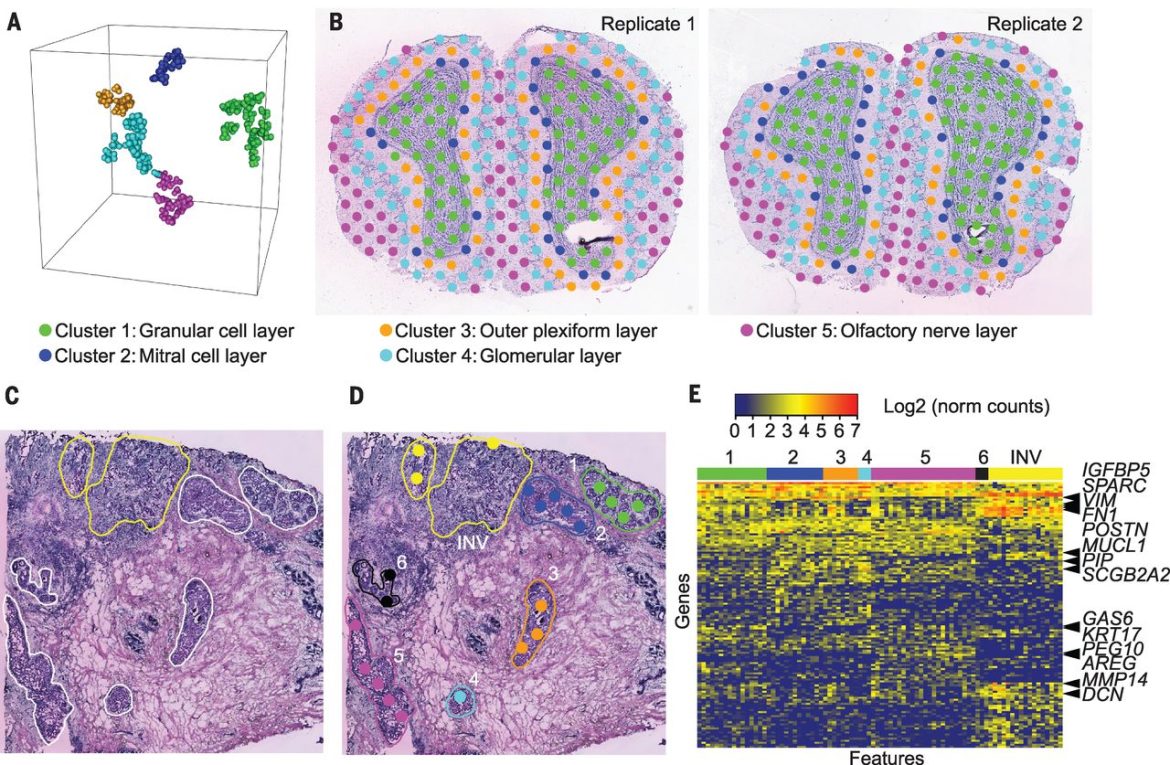
Spatial transcriptomics or spatial analysis is a technique that uses high-throughput sequencing to measure transcriptome changes in cells over time and space. This method is used to study cellular processes such as development, stress, and aging.
The 3-Step Process for Spatial Transcriptomic Analysis
The spatial transcriptomic analysis is a method of studying gene expression in different areas of the body. This is analyzing RNA molecules extracted from cells that are present in different areas of the body.
The 3-Step Process for Spatial Transcriptomic Analysis:
1) Gather samples: tissue samples is taken from any area of the body and frozen at -80 degrees Celsius for later use.
2) Extract RNA: using a specific extraction protocol, extract RNA molecules from cells present in the tissue sample.
3) Perform gene expression analysis: using bioinformatics tools, analyze spatial transcriptomic data to study the differences between cells’ gene expression levels across different areas of the body.
How Does It Work in Practice?
A bioinformatics software platform is a software that uses artificial intelligence to analyze, interpret and interpret data from biological experiments.
Bioinformatics software platforms have a variety of uses in scientific research. They use for gene expression analysis, transcriptomics, proteomics, and metabolomics.
A bioinformatics software platform is a computer-based system for analyzing and interpreting data from biological experiments. This type of software has a wide range of applications in scientific research, including gene expression analysis, transcriptomics, proteomics, and metabolomics.
What are the Key Considerations for a Successful Spatial Transcriptomic Analysis of a Patient’s Tissue Sections?
To understand the molecular mechanisms of disease, transcriptomic analysis is often used. The transcriptomic analysis is a technique that identifies the expression levels of transcripts in RNA samples. It is a powerful tool for understanding gene expression patterns and is used to identify potential biomarkers.
Key considerations for successful spatial transcriptomic analysis:
1) What are the data types?
2) What are the standards and quality control?
3) How much information do you need?
How do you figure out what genes are important to the process at hand?
The process of gene expression is a complex one. It’s not just about genes, but it also involves the environment, body size, and other factors. There are many ways that scientists can figure out what genes are important to the process at hand. One way is by looking at the differences in gene expression between two groups of organisms, like fruit flies and humans.
Scientists also look at how fast different genes are expressed in an organism’s cells. This is done by growing cells from an organism in a lab dish and then looking for proteins that are produced from those cells over time. The proteins is produced over time will show up as spots on an image taken of the cells over time.